| BRENDA, the digital lexicon of enzymes |
 |
| BRENDA, the digital lexicon of enzymes |
 |
Created and until today continuously being updated by Prof. Schomburg's expert team of molecular biologist (now at the University of Cologne) which combs through relevant biological literature for curation into BRENDA.
The curated data is presented in easy to use formats that offers enhanced flexibility and powerful time savings over traditional library research. Access to thousands of independent research results offers the competitive advantage of reduced library research time and faster progression through discovery pathways.
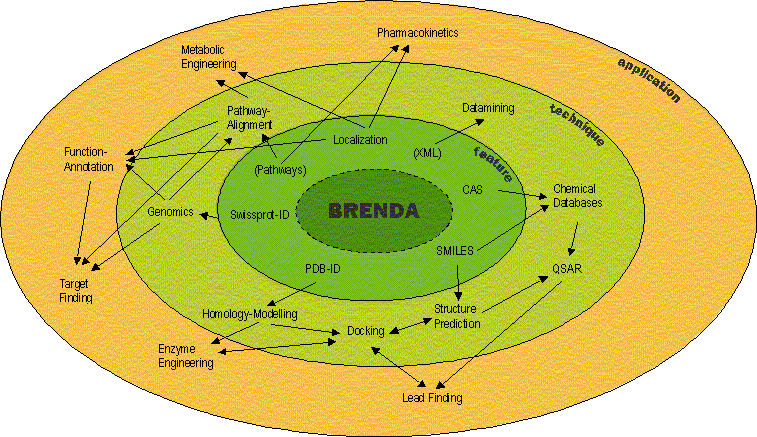
BRENDA covers a wide range of enzyme characteristics: More than 50 fields containing close to half a million entries can be queried in various combinations. All enzymes are characterized in an organism-specific manner, and all data are linked to primary literature references.
Within BRENDA inhibitors, cofactors, natural and artificial substrates are annotated with information about the molecular
structure in SMILES-Format (see Daylight, Inc. for information on SMILES).
For example see aminopterin:
Nc1nc(N)c2nc(CNc3ccc(cc3)C(=O)N[C@@H](CCC(=O)O)C(=O)O)cnc2n1 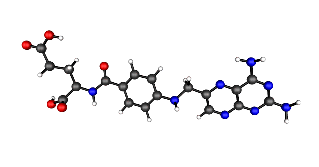
BRENDA augments your existing data with:
Current references to BRENDA :